
JSE Outstanding Papers
1. A framework infrageneric classification of Carex (Cyperaceae) and its organizing principles
(47 Citations in Web of Science Core Collection)
https://doi.org/10.1111/jse.12722

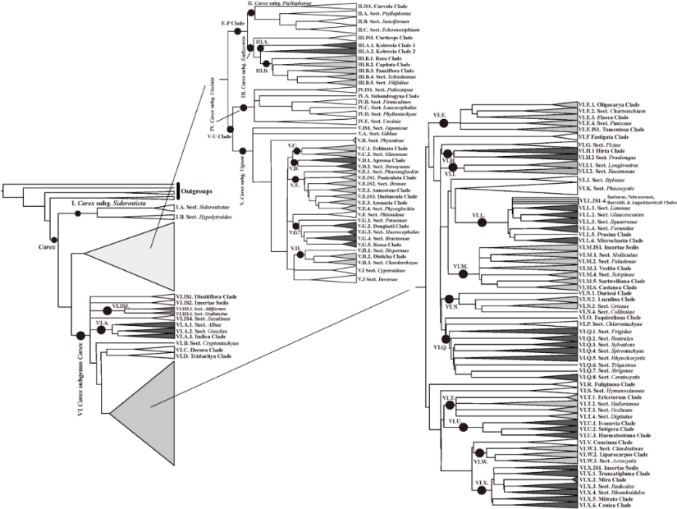
In this study, Roalson et al. (2021) presented a hybrid classification framework of Carex to reflect the current state of our knowledge using a combination of informally named clades and formally named infrageneric groups. Based on an order corresponding to a linear arrangement of the clades on a ladderized phylogeny, the authors organized Carex into six subgenera, which were further subdivided into 62 formally named sections plus 49 informal groups. As the authors remarked, as many as 113 species were not placed in a clade with any confidence in this study and difficulties remained for untangling intricate nomenclature and identifying types for the many available sectional names. Despite these challenges, the proposed infrageneric classification of Carex will serve as a roadmap for better organizing our understanding of phylogeny, identifying species groups and stimulating further studies of this important genus.
2. A new classification of Cyperaceae (Poales) supported by phylogenomic data
(37 Citations in Web of Science Core Collection)
https://doi.org/10.1111/jse.12757
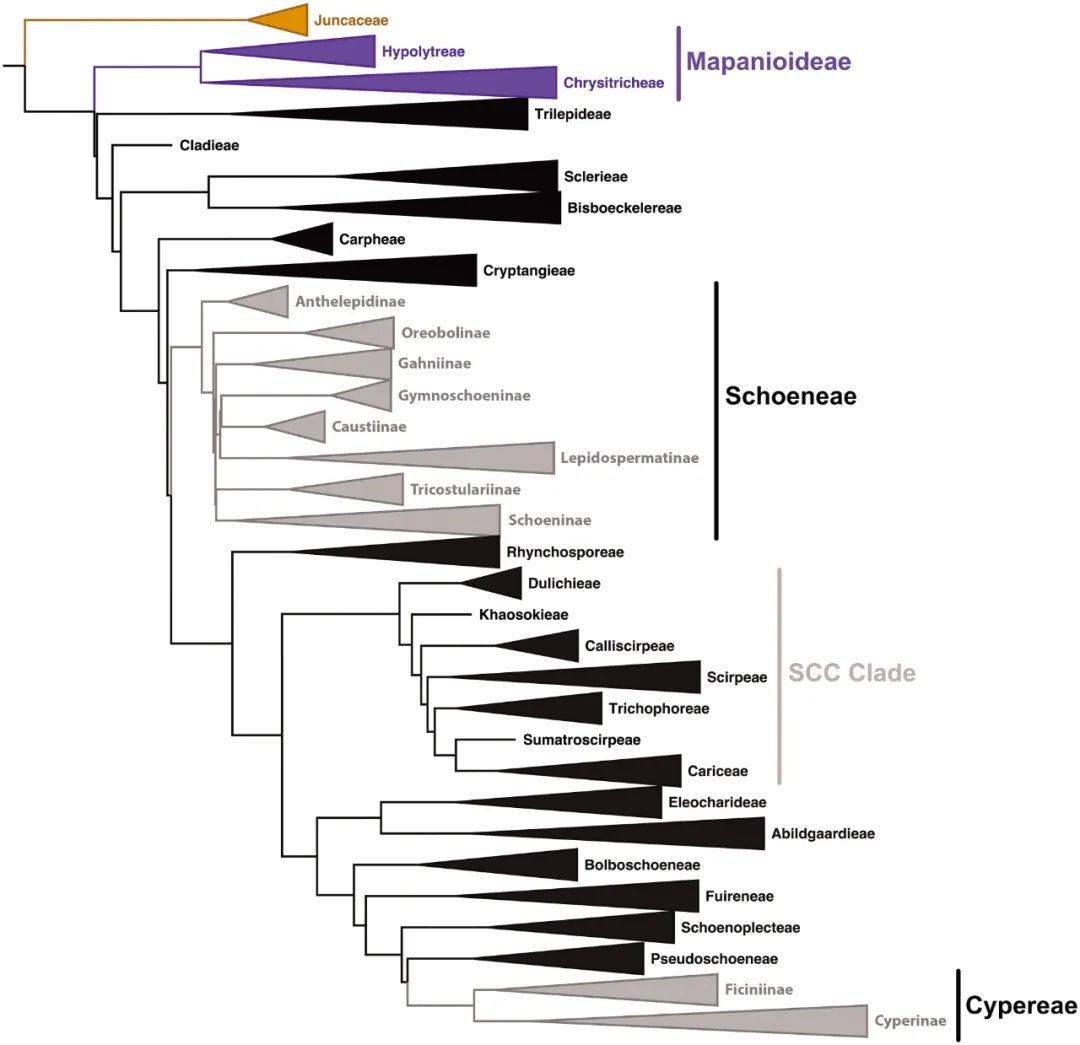
By sampling 361 accessions of Cyperaceae species and 21 accessions representing other families in the order Poales, Larridon et al. (2021) presented the first family‐wide phylogenomic study of Cyperaceae based on targeted sequencing using the Angiosperms probe kit. Their results supported the majority of previously recognized suprageneric groups, and placed previously unplaced genera in the family phylogeny for the first time. Based on the resolved phylogenetic framework, the authors proposed a new classification for the family at the tribal, subtribal, and generic levels, with a taxonomic treatment including identification keys and diagnoses for the two subfamilies, 24 tribes, and also 10 subtribes, and taxonomic information on the 95 genera recognized. Moreover, this study implicated that the high‐throughput technique—targeted sequencing is able to sequence historical herbarium specimens with poor DNA quality and thus might be widely applicable to molecular evolutionary studies in plants.
JSE Outstanding Papers by Young Investigators:
(37 Citations in Web of Science Core Collection)
https://doi.org/10.1111/jse.12585
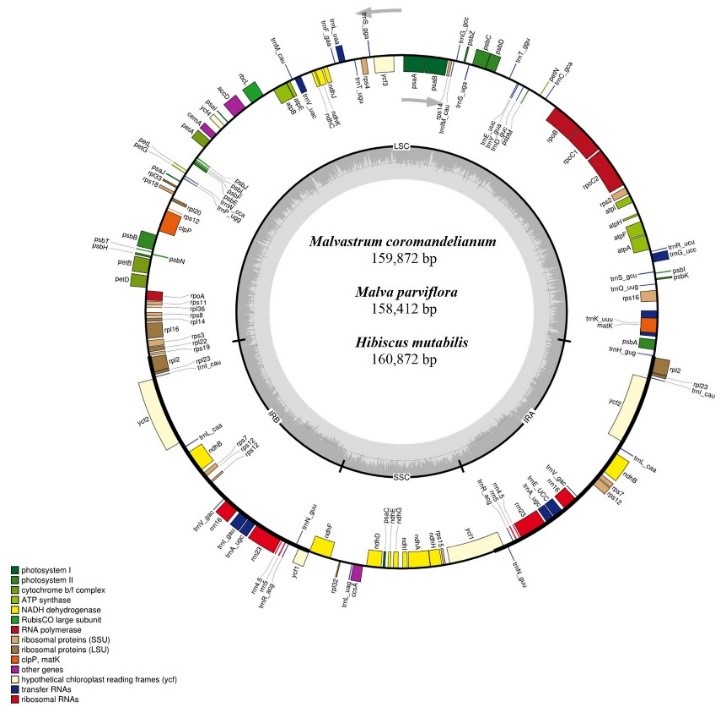
Based on 19 whole‐chloroplast genomes representing 12 genera of Malvaceae (three newly sequenced and 16 publicly available), Abdullah et al. (2021) calculated the correlation coefficients among these mutational events at family, subfamily, and genus levels. By defining a correlation coefficient to categorize the extent of correlations among substitutions, InDels and repeats, the authors revealed weak and strong correlations among these mutational events and confirmed the co‐occurrence of up to 90% of oligonucleotide repeats with substitutions. This study supported the previous argument that oligonucleotide repeats could be used as a proxy for finding the mutational hotspots. The authors also hypothesized that the correlations among these mutations might be a common characteristic of chloroplast genomes in all plant lineages, which deserves further investigations in various lineages of organisms.
2. Phylogeny and biogeography of the hollies (Ilex L., Aquifoliaceae)
(35 Citations in Web of Science Core Collection)
https://doi.org/10.1111/jse.12567

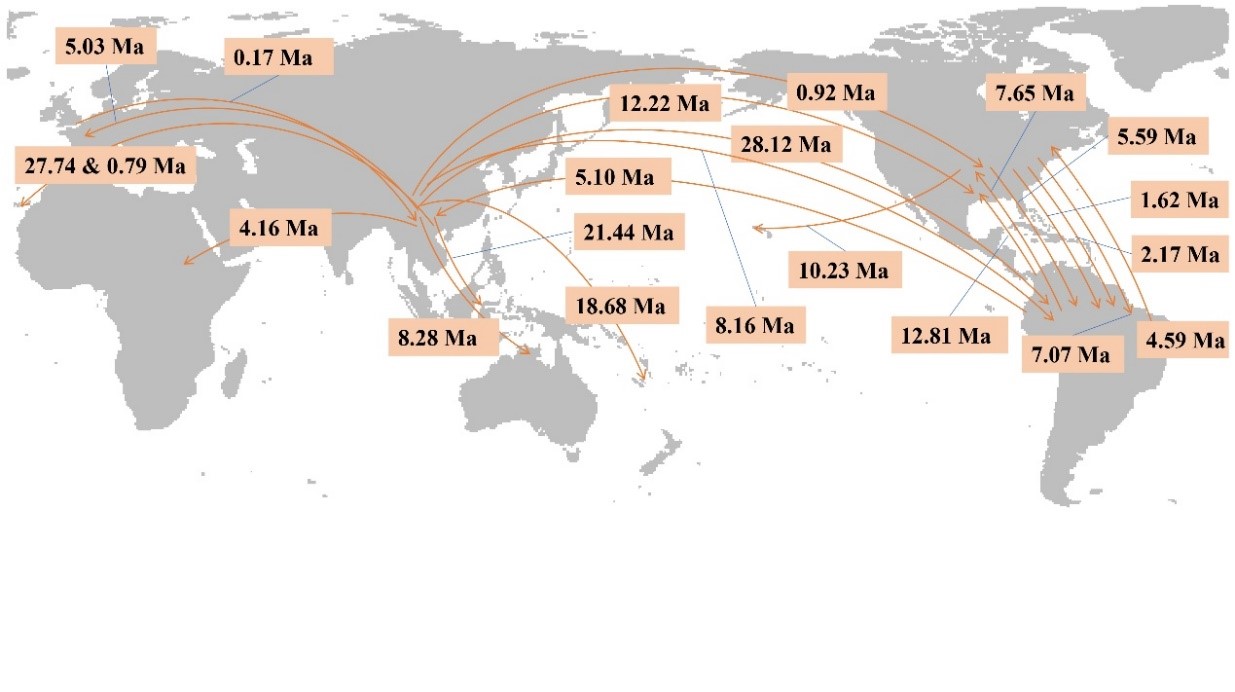
Based on sequences of two nuclear genes representing 177 species and calibrated using three macrofossil records, Yao et al. (2021) constructed a phylogeny of Ilex and investigated the biogeographic patterns of the genus. They identified five main clades that had a common ancestor in the early Eocene, much earlier than previously hypothesized. They also demonstrated that Ilex originated in subtropical Asia and subsequently colonized the Americas, Australia, Europe and Africa, and that the current near‐cosmopolitan distribution reflects a balance between dispersal, diversification and extinction. This work provides an important evolutionary framework for further studies in comparative ecology and evolution using Ilex as a feasible system.